Artificial Intelligence in Drug Discovery
Much of the existing hype in biotech has concentrated around the promise of revolutionising drug discovery. After all, the last decade was a so-called golden age in the field. From 2012 to 2021, compared to the prior decade, an increase of 73% new medicines were approved — 25% more than the one before that. These medicines include immunotherapies for cancer, gene therapies, and, of course, Covid vaccines. On the face of it, the pharmaceutical industry is doing well.
But there are increasingly worrying trends. Drug discovery is becoming prohibitively expensive and risky. As of today, it costs between $1bn-$3bn on average and 12–18 years to bring a new drug to market. Meanwhile, the average price of a new medicine has skyrocketed from $2k in 2007 to $180k in 2021.
That’s why many have pinned their hopes on Artificial Intelligence (AI) approaches (such as statistical machine learning) to help accelerate the development of new drugs, from early target identification through trials. While several compounds have already been identified using various machine learning algorithms, these are still in early discovery or preclinical stages of development. The promise of AI revolutionizing drug discovery is still a very exciting but unfulfilled promise.
What Is AI?
To fulfill that promise, it’s vital to understand what we really mean by AI. In recent years, the term has become quite the buzzword, devoid of much technical substance. So what actually constitutes Artificial Intelligence?
AI, as an academic field, has already been around since the 1950s, and over time, branched into various types to represent different ways of learning. These types are eloquently described in Professor Pedro Domingos’s book The Master Algorithm (he calls them “tribes”): connectionists, symbolists, evolutionaries, bayesians, and analogisers.
Over the past decade, bayesians and connectionists have received much of the public’s limelight, unlike the symbolists. Symbolists create true to life representations of the world based on sets of rules that make logical inferences. Symbolic AI systems do not have the enormous publicity that other types of AI enjoy, but they possess unique and important capabilities missing in others: automated reasoning and knowledge representation.
Representing Biomedical Knowledge
The issue of knowledge representation is, in fact, precisely one of the biggest problems in drug discovery. Existing database software, such as relational or graph databases, make it difficult to accurately represent and make sense of the immense intricacies and complexities of biology.
Drug discovery’s representation problem is well illustrated by the need to create unified models of disparate biomedical data sources (e.g., Uniprot or Disgenet). At the database level, this means creating data models (some may refer to these as ontologies) that describe a myriad of complex entities and relationships, such as those between proteins, genes, drug, diseases, interactions, and many others.
This is what TypeDB, an open-source database software, aims to achieve — to enable developers to create true to life representations of highly complex domains that computers can use to gain insights.
TypeDB’s type system is based on entity-relationship concepts that represents the data stored in TypeDB. This makes it expressive enough to accurately capture complex biomedical domain knowledge (through type inference, nested relations, hyper-relations, rule inference, and much more), making it easier for scientists to obtain insights and accelerate development timelines.
This was illustrated by a large pharmaceutical company that struggled for over five years to model a disease network using Semantic Web Standards, but managed to achieve this in just three weeks after migrating to TypeDB.
For example, a biomedical model written in TypeQL (TypeDB’s query
language) that describes proteins, genes, and diseases looks as follows:
define
protein sub entity,
owns uniprot-id,
plays protein-disease-association:protein,
plays encode:encoded-protein;
gene sub entity,
owns entrez-id,
plays gene-disease-association:gene,
plays encode:encoding-gene;
disease sub entity,
owns disease-name,
plays gene-disease-association:disease,
plays protein-disease-association:disease;
encode sub relation,
relates encoded-protein,
relates encoding-gene;
protein-disease-association sub relation,
relates protein,
relates disease;
gene-disease-association sub relation,
relates gene,
relates disease;
uniprot-id sub attribute, value string;
entrez-id sub attribute, value string;
disease-name sub attribute, value string;For a fully worked example, an open source biomedical knowledge graph can be found here on Github. This loads data from various well-known biomedical sources such as Uniprot, Disgenet, Reactome, and others.
With your data stored in TypeDB, you could run queries that ask questions such as:
Which drugs interact with genes associated to the virus SARS?
To answer that question, we could use the following query in TypeQL:
match
$virus isa virus, has virus-name "SARS";
$gene isa gene;
$drug isa drug;
($virus, $gene) isa gene-virus-association;
($gene, $drug) isa drug-gene-interaction; Running this will make TypeDB return the data that matches the conditions described in the query. This can be visualised in TypeDB Studio as follows, which will help to understand what relevant drugs may warrant further investigation.
Through automated reasoning, TypeDB can also infer knowledge that doesn’t exist in the database. This is done by writing rules, which form part of the schema in TypeDB. For example, a rule could infer an association between a gene and a disease, if the encoded protein by that gene is associated with that disease. Such a rule would be written as follows:
rule inference-example:
when {
(encoding-gene: $gene, encoded-protein: $protein) isa encode;
(protein: $protein, disease: $disease) isa protein-disease-association;
} then {
(gene: $gene, disease: $disease) isa gene-disease-association;
}; Then, if we were to insert the following data:
insert
$gene isa gene, has entrez-id "2";
$protein isa protein, has uniprot-id "P01023";
$disease isa disease, has disease-name "Parkinson Disease";
(encoding-gene: $gene, encoded-protein: $protein) isa encode;
(protein: $protein, disease: $disease) isa protein-disease-association;TypeDB would be able to infer a connection between the gene and the disease, even if none has been inserted into the database. In this case, the following relation gene-disease-association would be inferred:
match
$gene isa gene, has gene-id "2";
$disease isa disease, has disease-name $dn; ;
(gene: $gene, disease: $disease) isa gene-disease-assocation;Accelerating Target Discovery Through Machine Learning
With these (symbolic) representations of biomedical data in TypeDB, the contextual knowledge that is fed into machine learning algorithms can make them extremely powerful to uncover insights that, for example, can enable a drug discovery pipeline to find promising targets.
One approach for finding promising targets is to use link prediction algorithms. TypeDB’s rule engine allows such an ML model to learn based on facts inferred via reasoning. This means shifting from learning over flat, non-contextual data to learning over reasoned, contextual knowledge. One of the benefits is that predictions may be generalized beyond the scope of the training data according to the logical rules of the domain and reduce the quantity of training data required.
Such a drug discovery workflow would work as follows:
- Query TypeDB to create a subgraph full of contextual knowledge that leverages the full expressivity of TypeDB.
- Transform the subgraph into an embedding and ingest these into a graph learning algorithm.
- The predictions (e.g., as probability scores between gene-disease associations) can then be inserted into TypeDB and used to validate/prioritize certain targets.
With those predictions in the database, we can ask higher level questions that leverage these predictions with the wider contextual knowledge in the database. For example:
What are the most likely gene targets for Melanoma that encode for proteins expressed in melanocytes?
Written in TypeQL, this question would look as follows:
match
$gene isa gene, has gene-id $gene-id;
$protein isa protein;
$cell isa cell, has cell-type "melanocytes";
$disease isa disease, has disease-name "melanoma";
($gene, $protein) isa encode;
($protein, $cell) isa expression;
($gene, $disease) isa gene-disease-association, has prob $p;
get $gene-id; sort desc $p;The result of this query would be a list of genes sorted by a probability score (as predicted by the graph learner):
{$gid "TOPGENE" isa gene-id;}
{$gid "BESTGENE" isa gene-id;}
{$gid "OTHERTARGET" isa gene-id;}
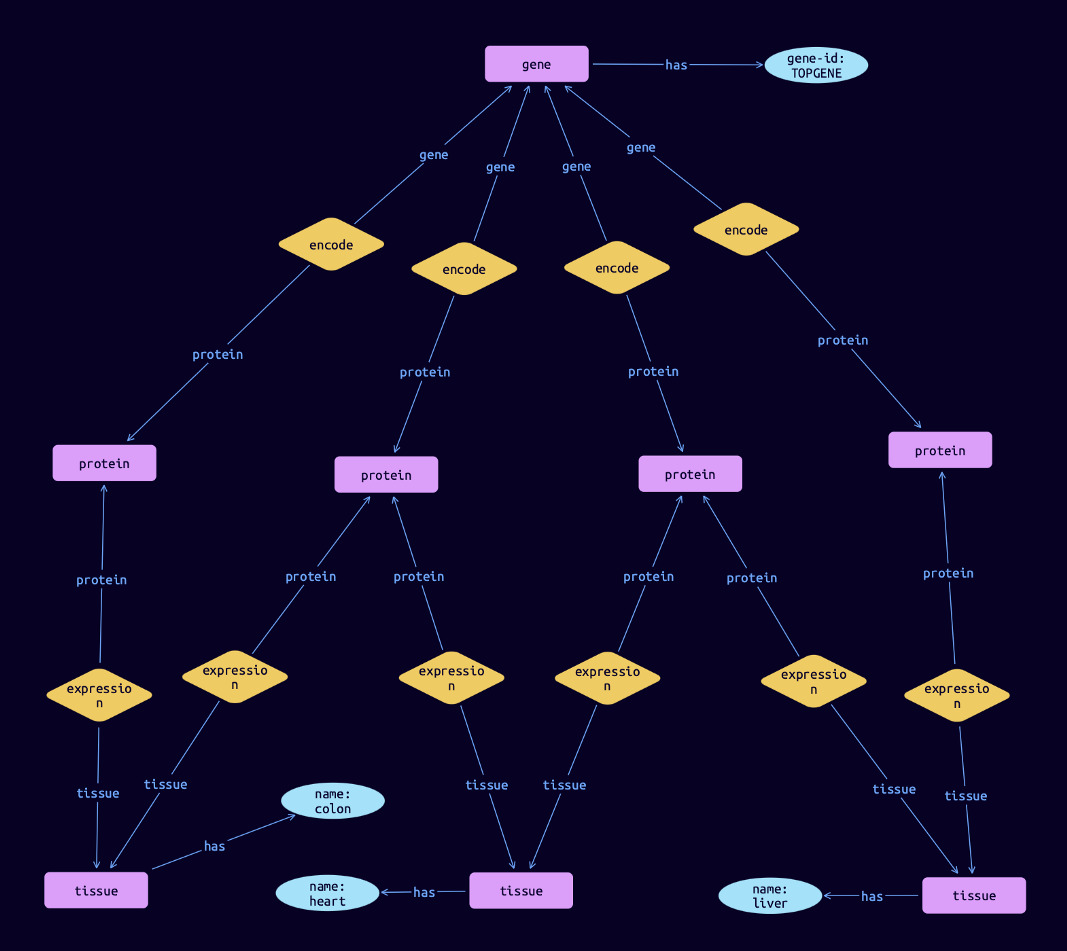
...We could then further investigate these genes, for example by understanding the biological context of each one. Let’s say we want to know the tissues in which the TOPGENE gene encodes proteins for. We could write the following query:
match
$gene isa gene, has gene-id $gene-id; $gene-id "TOPGENE";
$protein isa protein;
$tissue isa tissue, has name $name;
$rel1 ($gene, $protein);
$rel2 ($protein, $tissue);The result, visualised in TypeDB Studio, could show this gene encoding for proteins that are expressed in the colon, heart, and liver:
Conclusion
The world urgently needs solutions to create treatments for devastating diseases and the hope is that, through innovations in AI, we can build a healthier world in which no disease goes untreated. The potential for AI to revolutionise drug discovery is still in its infancy, but fulfilling that promise will see biology unleash a new wave of innovation and make the 21st truly its century.
In this article, we’ve looked at how TypeDB contributes to that journey by enabling the symbolic representation of biomedical knowledge and improving statistical ML with it. Scientists applying AI in drug discovery use TypeDB to analyse disease networks, get a better understanding of the ever growing complexities of biomedical research, and uncover new and ground breaking treatments.
If you’re looking for an example of how to aggregate biomedical data in TypeDB in a way that is leverageable for drug discovery, have a look at this useful resource: TypeDB Bio.
Much of the existing hype in biotech has concentrated around the promise of revolutionising drug discovery. After all, the last decade was a so-called golden age in the field. From 2012 to 2021, compared to the prior decade, an increase of 73% new medicines were approved — 25% more than the one before that. These medicines include immunotherapies for cancer, gene therapies, and, of course, Covid vaccines. On the face of it, the pharmaceutical industry is doing well.
But there are increasingly worrying trends. Drug discovery is becoming prohibitively expensive and risky. As of today, it costs between $1bn-$3bn on average and 12–18 years to bring a new drug to market. Meanwhile, the average price of a new medicine has skyrocketed from $2k in 2007 to $180k in 2021.
That’s why many have pinned their hopes on Artificial Intelligence (AI) approaches (such as statistical machine learning) to help accelerate the development of new drugs, from early target identification through trials. While several compounds have already been identified using various machine learning algorithms, these are still in early discovery or preclinical stages of development. The promise of AI revolutionizing drug discovery is still a very exciting but unfulfilled promise.
What Is AI?
To fulfill that promise, it’s vital to understand what we really mean by AI. In recent years, the term has become quite the buzzword, devoid of much technical substance. So what actually constitutes Artificial Intelligence?
AI, as an academic field, has already been around since the 1950s, and over time, branched into various types to represent different ways of learning. These types are eloquently described in Professor Pedro Domingos’s book The Master Algorithm (he calls them “tribes”): connectionists, symbolists, evolutionaries, bayesians, and analogisers.
Over the past decade, bayesians and connectionists have received much of the public’s limelight, unlike the symbolists. Symbolists create true to life representations of the world based on sets of rules that make logical inferences. Symbolic AI systems do not have the enormous publicity that other types of AI enjoy, but they possess unique and important capabilities missing in others: automated reasoning and knowledge representation.
Representing Biomedical Knowledge
The issue of knowledge representation is, in fact, precisely one of the biggest problems in drug discovery. Existing database software, such as relational or graph databases, make it difficult to accurately represent and make sense of the immense intricacies and complexities of biology.
Drug discovery’s representation problem is well illustrated by the need to create unified models of disparate biomedical data sources (e.g., Uniprot or Disgenet). At the database level, this means creating data models (some may refer to these as ontologies) that describe a myriad of complex entities and relationships, such as those between proteins, genes, drug, diseases, interactions, and many others.
This is what TypeDB, an open-source database software, aims to achieve — to enable developers to create true to life representations of highly complex domains that computers can use to gain insights.
TypeDB’s type system is based on entity-relationship concepts that represents the data stored in TypeDB. This makes it expressive enough to accurately capture complex biomedical domain knowledge (through type inference, nested relations, hyper-relations, rule inference, and much more), making it easier for scientists to obtain insights and accelerate development timelines.
This was illustrated by a large pharmaceutical company that struggled for over five years to model a disease network using Semantic Web Standards, but managed to achieve this in just three weeks after migrating to TypeDB.
For example, a biomedical model written in TypeQL (TypeDB’s query
language) that describes proteins, genes, and diseases looks as follows:
define
protein sub entity,
owns uniprot-id,
plays protein-disease-association:protein,
plays encode:encoded-protein;
gene sub entity,
owns entrez-id,
plays gene-disease-association:gene,
plays encode:encoding-gene;
disease sub entity,
owns disease-name,
plays gene-disease-association:disease,
plays protein-disease-association:disease;
encode sub relation,
relates encoded-protein,
relates encoding-gene;
protein-disease-association sub relation,
relates protein,
relates disease;
gene-disease-association sub relation,
relates gene,
relates disease;
uniprot-id sub attribute, value string;
entrez-id sub attribute, value string;
disease-name sub attribute, value string;For a fully worked example, an open source biomedical knowledge graph can be found here on Github. This loads data from various well-known biomedical sources such as Uniprot, Disgenet, Reactome, and others.
With your data stored in TypeDB, you could run queries that ask questions such as:
Which drugs interact with genes associated to the virus SARS?
To answer that question, we could use the following query in TypeQL:
match
$virus isa virus, has virus-name "SARS";
$gene isa gene;
$drug isa drug;
($virus, $gene) isa gene-virus-association;
($gene, $drug) isa drug-gene-interaction; Running this will make TypeDB return the data that matches the conditions described in the query. This can be visualised in TypeDB Studio as follows, which will help to understand what relevant drugs may warrant further investigation.
Through automated reasoning, TypeDB can also infer knowledge that doesn’t exist in the database. This is done by writing rules, which form part of the schema in TypeDB. For example, a rule could infer an association between a gene and a disease, if the encoded protein by that gene is associated with that disease. Such a rule would be written as follows:
rule inference-example:
when {
(encoding-gene: $gene, encoded-protein: $protein) isa encode;
(protein: $protein, disease: $disease) isa protein-disease-association;
} then {
(gene: $gene, disease: $disease) isa gene-disease-association;
}; Then, if we were to insert the following data:
insert
$gene isa gene, has entrez-id "2";
$protein isa protein, has uniprot-id "P01023";
$disease isa disease, has disease-name "Parkinson Disease";
(encoding-gene: $gene, encoded-protein: $protein) isa encode;
(protein: $protein, disease: $disease) isa protein-disease-association;TypeDB would be able to infer a connection between the gene and the disease, even if none has been inserted into the database. In this case, the following relation gene-disease-association would be inferred:
match
$gene isa gene, has gene-id "2";
$disease isa disease, has disease-name $dn; ;
(gene: $gene, disease: $disease) isa gene-disease-assocation;Accelerating Target Discovery Through Machine Learning
With these (symbolic) representations of biomedical data in TypeDB, the contextual knowledge that is fed into machine learning algorithms can make them extremely powerful to uncover insights that, for example, can enable a drug discovery pipeline to find promising targets.
One approach for finding promising targets is to use link prediction algorithms. TypeDB’s rule engine allows such an ML model to learn based on facts inferred via reasoning. This means shifting from learning over flat, non-contextual data to learning over reasoned, contextual knowledge. One of the benefits is that predictions may be generalized beyond the scope of the training data according to the logical rules of the domain and reduce the quantity of training data required.
Such a drug discovery workflow would work as follows:
- Query TypeDB to create a subgraph full of contextual knowledge that leverages the full expressivity of TypeDB.
- Transform the subgraph into an embedding and ingest these into a graph learning algorithm.
- The predictions (e.g., as probability scores between gene-disease associations) can then be inserted into TypeDB and used to validate/prioritize certain targets.
With those predictions in the database, we can ask higher level questions that leverage these predictions with the wider contextual knowledge in the database. For example:
What are the most likely gene targets for Melanoma that encode for proteins expressed in melanocytes?
Written in TypeQL, this question would look as follows:
match
$gene isa gene, has gene-id $gene-id;
$protein isa protein;
$cell isa cell, has cell-type "melanocytes";
$disease isa disease, has disease-name "melanoma";
($gene, $protein) isa encode;
($protein, $cell) isa expression;
($gene, $disease) isa gene-disease-association, has prob $p;
get $gene-id; sort desc $p;The result of this query would be a list of genes sorted by a probability score (as predicted by the graph learner):
{$gid "TOPGENE" isa gene-id;}
{$gid "BESTGENE" isa gene-id;}
{$gid "OTHERTARGET" isa gene-id;}
...We could then further investigate these genes, for example by understanding the biological context of each one. Let’s say we want to know the tissues in which the TOPGENE gene encodes proteins for. We could write the following query:
match
$gene isa gene, has gene-id $gene-id; $gene-id "TOPGENE";
$protein isa protein;
$tissue isa tissue, has name $name;
$rel1 ($gene, $protein);
$rel2 ($protein, $tissue);The result, visualised in TypeDB Studio, could show this gene encoding for proteins that are expressed in the colon, heart, and liver:
Conclusion
The world urgently needs solutions to create treatments for devastating diseases and the hope is that, through innovations in AI, we can build a healthier world in which no disease goes untreated. The potential for AI to revolutionise drug discovery is still in its infancy, but fulfilling that promise will see biology unleash a new wave of innovation and make the 21st truly its century.
In this article, we’ve looked at how TypeDB contributes to that journey by enabling the symbolic representation of biomedical knowledge and improving statistical ML with it. Scientists applying AI in drug discovery use TypeDB to analyse disease networks, get a better understanding of the ever growing complexities of biomedical research, and uncover new and ground breaking treatments.
If you’re looking for an example of how to aggregate biomedical data in TypeDB in a way that is leverageable for drug discovery, have a look at this useful resource: TypeDB Bio.
